RNA later and 2D SDS PAGE - (Mar/07/2016 )
Hi
I'm wondering does anyone have any experience or advice on using tissue that has been stored in RNAlater for 2D SDS PAGE?
I'm having a lot of problems with it. The tissue has been stored at -20C in RNAlater. I have extracted in 2D lysis buffer (urea/thiourea/chaps/dtt) and then dialysed against dH2O followed by acetone precipitation and rehydration of the pellet in 2D buffer followed by centrifugation.
On a 1D gel there are no distinct bands present, it's just more of a smear. On a 2D gel, there are lots of horizontal streaks.
Has anyone had any experience with this?
thanks
i don't have experience with rnalater but it sounds like you haven't completely solubilized your precipitated proteins.
Hi
Thanks for your reply. I solubilized in urea/thiourea/chaps/dtt until the pellet is completely dissolved. Then spun at 15,000 rpm to remove any insoluble material. I am left with a clear supernatant. What else can I do to make sure that the protein has been completely solubilized?
thanks
I've attached an image of the 1D gel. The first lane is the rainbow markers and the second lane is the sample in question.
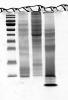
your gel doesn't look as bad as you said. you do see distinct bands, not smears, they are not as sharp as you may have hoped but that may be due to such factors as complexity of extract, loading volume, amount of protein loaded, time between stopping run and fixation, solubilization solution (detergents will affect migration and the gel, decomposing urea will carbamylate the proteins), residual organics (acetone) will affect the gel, to name a few.
streaks in the 2nd dimension are often due to insufficient denaturing after focusing.
