EMSA: cell extract vs purified protein - (Feb/08/2013 )
Hi everyone,
I am planning to perform EMSA with a supposedly DNA binding protein and a 22-mer oligo DNA. But I was wondering what's the difference between using purified protein fraction versus cell extract extracts + noncompetitve DNA such as dIdC?
My protein is transiently transfected to human cells using lipofectamine 2000. I can already have my protein of interest purified from 293T human cells using anti-FLAG beads, but I can only get this much (see attached picture) protein. It's also time consuming and costly. I already tried to use the purified protein to do EMSA several times and I get little to no binding.
I'm reluctant to use the cell lysate because the expression may not be enough. Is this much protein expression enough for whole cell EMSA?
Thanks
qp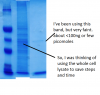
whole cell lysate can give you a lot of non-specific binding. I think you can try expressing your protein in bacteria and use the purified protein for EMSA or use nuclear extract from HeLa cells. For proteins to bind DNA, it must be nuclear so nuclear extract is typically used for EMSA.
pcrman on Sat Feb 9 02:02:12 2013 said:
whole cell lysate can give you a lot of non-specific binding. I think you can try expressing your protein in bacteria and use the purified protein for EMSA or use nuclear extract from HeLa cells. For proteins to bind DNA, it must be nuclear so nuclear extract is typically used for EMSA.
I was wondering if the DNA-protein binding can be improved by using the whole cell lysate of my human cells instead of purified proteins. Of course, I will include a negative control mutant DNA that's not supposed to bind to my protein of interest.
