ChIP sonication - (Oct/03/2012 )
Hi
I have been trying to optimize sonication of the chromatin from rat brain for ChIP. Please find below the gel image of the decrosslinked, proteinase K treated and phenol washed DNA. Sonication was done with Bioruptor (1) 5 (2) 10 (3) 15 and (4) 20 cycles of 30 sec on/off at high power. Does any of these chromatin samples appears to be of the quality of being taken ahead for immunoprecipitation? I can provide details of steps performed after tissue harvest and before sonication.
Thank you in anticipation

Lane 3 and 4 have a nice distribution of the fragments, but seem a bit over sonicated. Line 2 is not enough.
Hi,
Is your problem solved? I have some problems with my ChIP assay. Hope you can give me some suggestions.

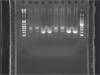
I did not do any experiment after above attempt.
What's the marker size in your case? I have consistently observed that if u load the sonicated samples directly (without phenol purifyling and having 1% SDS coming from lysis buffer in my case) onto the agarose gel, you get some weared pattern. I am sure you have done the same. So my suggesiton is that after RNase-proteinaseK incubation give phenol-chloroform treatment followed by alcohol (isopropanol or ethanol) precipitation and than load the samples on gel for analyzing the size.
Hi rmbio, the marker in the fist lane is 1Kb Plus DNA ladder and in the last lane is 100bp DNA ladder. I will try the phenol-chloroform treatment. Thank you very much for your suggestion!
