SDM - (Apr/30/2009 )
I have been doing site directed mutagenesis in the past few months.. hoping to create single amino acid mutant for alpha synuclein. I did the PCR, dpnI digestion and transformation..changing some conditions here and there..and FINALLY i got a lot of colonies on my LB+ Amp plate.
But i'm using self prepared supercompetent DH5alpha cells to do the transformation.
After plasmid extraction, i run an agarose gel analysis and found out that other than the band(5kb) that i wanted..there are few more bands which i don't really know what are they..I'm worried of contamination with the supercompetent cells.
I have attached the gel pic taken..and I'm hoping that anyone who know can help me with this..
thanks in advance..^^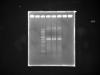
Hey,
1. Ladder and the lanes are not marked.
2. Is it digested with a restriction enzyme? If not then you expect multiple bands. However I would suggest that you digest with some single cutter and make sure that the size is fine.
3. I generally introduce or delete some restriction site in the SDM reaction to quickly check for mutagenesis by digestion alone.
Hope it helps.
Best,
TC
chijing on Apr 30 2009, 12:32 PM said:
But i'm using self prepared supercompetent DH5alpha cells to do the transformation.
After plasmid extraction, i run an agarose gel analysis and found out that other than the band(5kb) that i wanted..there are few more bands which i don't really know what are they..I'm worried of contamination with the supercompetent cells.
I have attached the gel pic taken..and I'm hoping that anyone who know can help me with this..
thanks in advance..^^
Oops..i'm so sorry..
For the gel pic attached:
1 lane-empty
2 lane-empty
3 lane-1kb DNA ladder
4 lane-miniprep plasmid DNA (i)
5 lane-miniprep plasmid DNA (ii)
6 lane-miniprep plasmid DNA (iii)
7 lane-empty
8 lane-empty
My expected band size should be 4.4kb(vector backbone)+422bp(insert)..around 4.8kb..
those plasmid DNA run on gel weren't digested..
I tried digesting the plasmid DNA before..but it seemed to show no changes btw the be4 and after digestion..
i usually digest the plasmid DNA with 1 ul RE at 37'C for around 2 hours? will it be long enuf for the digestion to occur?
T C on Apr 30 2009, 04:34 PM said:
1. Ladder and the lanes are not marked.
2. Is it digested with a restriction enzyme? If not then you expect multiple bands. However I would suggest that you digest with some single cutter and make sure that the size is fine.
3. I generally introduce or delete some restriction site in the SDM reaction to quickly check for mutagenesis by digestion alone.
Hope it helps.
Best,
TC
chijing on Apr 30 2009, 12:32 PM said:
But i'm using self prepared supercompetent DH5alpha cells to do the transformation.
After plasmid extraction, i run an agarose gel analysis and found out that other than the band(5kb) that i wanted..there are few more bands which i don't really know what are they..I'm worried of contamination with the supercompetent cells.
I have attached the gel pic taken..and I'm hoping that anyone who know can help me with this..
thanks in advance..^^
![]()
I could make out the lanes but how do I read the ladder? what size is the thick band at?
Besides it doesn't look that the digestion is happening. It may sound stupid of me asking this but hope that the restriction site is there on the plasmid. Also, make sure that the restriction enzyme is working. Digest with another enzyme, run on the gel and check. As a control for checking if the enzyme is working, you cn use the template that you used for digestions.
It depends upon the enzyme but generally two hrs of digestion is enough.
I personally think taht teh enzyme is the ssue and you have yr SDM reaction working as after DpnI digestion you have a good band.
Best,
TC
chijing on Apr 30 2009, 06:00 PM said:
those plasmid DNA run on gel weren't digested..
I tried digesting the plasmid DNA before..but it seemed to show no changes btw the be4 and after digestion..
i usually digest the plasmid DNA with 1 ul RE at 37'C for around 2 hours? will it be long enuf for the digestion to occur?
