Question regarding basic trick in R - (Dec/12/2018 )
Dear All,
This is a basic question in R but I have been for a long time trying to catch the trick, but could not.
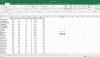
Every time when I import CSV files (table) into R using the set directory function and then read.csv function and giving it a name, I obtain a data frame, but the row names were placed among variables as column 1 and thus the column 0 is just the series of numbers 1, 2, 3, 4, etc set by default in R.
What I want is to obtain the table, but the row names should be given column 0 and the first variable is column 1, then if I am making multidimentional scaling, I can plot the names of the row rather than the numbers 1, 2, 3, etc,
How can I do that ? is this because of a different exportation function? I am sure that my table as attached contain the first column as row names and the first row as column names.
Thanks in advance
Are you viewing your data in the console/terminal using something like "head(SS)" or even just "SS" ? If so, R appends row numbers to the file. These are not a column, just representation of the individual rows, just as they are in Excel (look to the left of your data in the picture you provided), and are not part of the "spreadsheet" as such. If you are using Rstudio (I strongly recommend it), then you can use it to view the spreadsheet in a more familiar way - type: view(SS)
Row names are variables - there is no getting around this. You can plot the names on the plot without too much trouble - try using the package ggfortify. You can also convert the results (matrix) into a data frame so you can use things like ggplot by wrapping the cmdscale command in data.frame: ss_dataframe <- data.frame(cmdscale(yourdata))
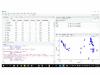
You are right. However, viewing them as View(SS), it gives the same as I told previously. You are correct in saying that it is just number. If this the case, why it gave me when making a MDS plot, just the numers (instead of the row names) in the plot ?? see attached as you see:
1. I created a distance using:
mds <- SS%>%
dist() %>%cmdscale() %>%
as_tibble()colnames(mds) <- c("Dim.1", "Dim.2")
Than when I plot the MDS, by
ggscatter(mds, x = "Dim.1", y = "Dim.2",
label = rownames(swiss),
size = 1, repel = TRUE)
It gives the numbers as a name for the row names but not the subjects themselves ?
Also when I run head (SS), it counts the row names as a variable and even this clear from the data view beside the name of the data set look at the upper right corner of the attached photo
I see what you mean, however the column 0 is not really a column (as I understand it, I could easily be completely wrong, being new to R myself). I found a few answers using google: https://support.bioconductor.org/p/79069/ seems like a good one.
In case you are not aware of it, stackoverflow.com is an excellent resource for any programming answers - R is pretty heavily questioned on there.\
I have no experience with MDS, but i think in your last call to label you (may) need "row.names" instead of "rownames". It's also possible that the ggscatter can't find swiss if it is a separate tibble - you might need to do a mutate to add the rownames from `swiss` to `mds`
