restriction enzymes/sites to avoid - (Apr/11/2014 )
Dear all,
I can (vaguely) remember reading something about restriction enzymes that you should avoid when doing cloning.
Too bad, I forgot what exactly it was and I also cant remember the enzymes..
Anyone an idea what restriction enzymes/sites one should avoid for cloning?
If you are using a manufacturer provided plasmid, chances are each RE present in the multiple cloning site has been thoroughly tested and will give decent results. I tend to avoid RE sites that produce weird overhangs or require any (N) nucleotide to cut. Always avoid compatible end producing enzymes (isoschizomers)
SalI has a bad reputation when used as a site created by placing it on the 5' end of a PCR primer.
I always choose enzymes which can be heat killed, which often avoids a purification step.
Choose cheap enzymes.
SalII thats it!
I remember this. But why would it cause problems ?
I often dont even do a purification step.. Even heat inactivation is something I often dont do when transforming cells. So far I had no problems with this.
@jerryshelly,
what do you mean by "weird overhangs" ?
And require any nucleotide to cut? You mean the standard "rule" they often mention that you need some extra bps to have it cut your restriction site?
Isoschizomers, I can see this also as an advantage , but I guess this depends on the reason why you use it.
Weird overhangs such as AleI that produce weird cut sites with "any" nucleotide. I just try to avoid using these all together.
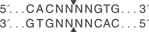
Edit - I guess they are not necessarily weird, I just prefer not to tinker with them. I like my common REs.
