Manipulation of target DNA fragments - (Nov/13/2013 )
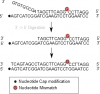
Hello, I have an experiment were I am capturing a target (single stranded DNA) and need to adapt/remove it's 3' overhang, so that the targets 3' now will match the 5' region of the probe (See picture). Additionally, the target might also contain a mismatch of interest (See picture), which should be conserved, so I believe DNA polymerases with 3'->5' proof reading is not an option in this experiment.
I have been considering the Mung bean nuclease which is capable of removing single stranded DNA from the 3' and 5' ends and then afterwards add DNA polymerase (without proof reading) to fill in the strand. However, I am uncertain whether the digested product from the Mung bean digestion can be elongated by a DNA polymerase afterwards so that the targets 3' area matches the probes 5'.
I hope you can answer my question or alternatively suggest other enzymes or approaches to achieve my goal
Best Regards
Heinz
AFAIK for the elongation of the 3' end, you need only the OH group intact. MBN nuclease generates ligable fragments, that would mean intack OH group on 3' end and phospho group on 5'.
