Anyone else see GFP alone normally localize at the centrosome? - (Nov/12/2013 )
Hello all,
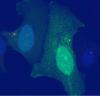
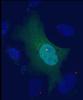
I'm trying to prove that my protein of interest colocalizes with the centrosome in various ovarian cancer cells (SKOV3, HEY, OVCA). I fused my protein of interest with GFP and did a transient transfection to introduce the construct into each of these cells. I also introduced constructs with only GFP into a separate group of cells as a control. I fixed the cells with 4% PFA and used a pericentrin antibody to confirm any colocalization. The typical GFP signal is seen but, strangely enough, the construct with the GFP alone also tends to develop a very bright structure that colocalizes with the centrosome. As far as I know, this should not be happening. Other research with centrosomes using a similar setup don't show any colocalization between GFP alone and the centrosome. I also see the colocalization with the GFP fusion protein, but it is not as strong as the constructs with the GFP alone. Given the colocalization with the GFP alone construct, I can't really conclude anything regarding colocalization of the centrosome and my protein of interest.
Anyone have any ideas why this could be happening/ how I can stop this from occurring?
Thanks to all.
GFP is normally seen throughout the cell, pretty evenly distributed. I would check that the GFP only construct you have, is actually what you think it is (sequence the GFP region), and try with a different GFP only construct too, to see if there is some difference in the signal.
For the most part, the GFP signal looks evenly distributed within the cell, but in my unfortunate case, it also has that bright signal at the centrosome. I don't currently have on me another GFP-only construct, but I did try a GFP-HA construct as well. Unfortunately, I also still see the same phenomenon. Is there any other cause that could be leading to this issue, other than an incorrect GFP sequence? I have attached an image of the GFP-only and the GFP-HA cells.
Thanks for all your help once again.
No idea sorry, it may not be due to the GFP sequence itself, but rather that your empty plasmid isn't actually empty, though this gets more unlikely as you use a range of constructs.
Is it possible that maybe my fixation procedure could be producing this artifact? I've been fixing my cells in 4% PFA for 12-15 minutes. They're later permeabilized in 0.1% TX and 0.02% BSA for the addition of the centrosome antibody after blocking in 8% BSA. I've been looking around, but I get conflicting answers. Some say methanol will bleach my GFP and others say GFP and methanol are a match made in heaven. ![]()
I've been using methanol fix with no problems with GFP, so I guess you could try it. PFA is the standard fix for these sorts of things though, and I would be very surprised if it is causing this artifact, unless there was some auto-fluorescence going on (which you would see in GFP negative cells). You could try reducing the PFA strength (I use 2% for 10 min) and see how that goes.
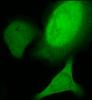
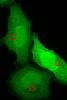
So I decided I'd try to at least pinpoint whether the issue was occurring due to the transfection itself or due to the fixation procedure. I took 2 groups of cells, transfected with the GFP-Only construct. One group had a MeOH fixation, the other PFA. I imaged them before and after fixation. Pictures are below. The cells prior to fixation had no signal that seemed to correspond to the centrosomes. After the PFA fixation though, a perinuclear signal appeared. Methanol fixation seemed to bleach my GFP signal completely. My plan is to try the lower concentration of PFA my next go around, but is there any other reliable fixation alternative that doesn't use PFA or methanol?
Thanks!
Fixatives for IF are usually grouped into two types - aldehydes (e.g. PFA, glutaraldehyde) or precipitating (alcohols), but there are a number of others that work. I know someone who is using zinc salts as a fixative, but I don't have her protocol about.
Your best bet would be to talk to a histologist or find a good histology text (try current protocols).
The 'spots' of GFP wouldn't be ribosomal would they? I've seen high concentrations of newly synthesized GFP-tagged proteins that is ribosomal in origin. WHen I pre-treat cells with cyclohexamide to stop protein synthesis the spots are no longer there.
No clue if they're ribosomal but anything is possible at this point with my cells. How long do you usually treat your cells with cyclohexamide to stop any protein synthesis?