Are these primer products good enough for qPCR? - (May/14/2013 )
Hello All,
Please review the attached photo and let me know if the first three primers are good enough for qPCR (PECAM-1, VEcad, and ACTA2). I designed these primers on NCBI's primerBlast and also enabled the option to search for unintended targets, in which only one target returned (no unintended targets).
The picture below the PCR was ran at 35 cycles with a 1 min. extension time. My amplicons are less than 200 bp though and for qPCR my extension time will be 30 seconds. The point I'm trying to get at is whether the products seen below 172 for PECAM-1, around 600 for VEcad, and around 800 for ACTA2 are from adjustable parameters in the PCR or from poor primer design.
Thank you for any insight. Some details: 10 ng cDNA Template, 0.2 uM For & Rev primer concentration
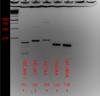
Are their other isoforms, pseudogenes? I like my qPCR products to give one sharp precise band. I would try redesigning my primers or adjust the cycling conditions. You could probably get those bands to disappear. Do not use 18S rRNA as a control. Find a more valid control for publication.
Edit: If you are using software to measure the relative density of each band, it may be alright. You could get a rough estimate, but I would personally try altering your methods first.
Thanks jerryshelly1 for your reply. I went ahead and ran qPCR with a 15 sec elongation time, since the amplicons were less than 250 bps (if 1 min = 1000 bp then 30 sec = 500 bp and 15 sec = 250 bp) then confirmed the quality of the data by running the qPCR products on a 2% agarose gel. I believe the bands that are present are of good enough quality for a proper qPCR. Thanks for your help again.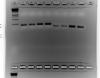
Looks beautiful. Good luck
