Cause of random samples failing PCR? - (May/03/2013 )
When running various PCR reactions I get random samples that fail while other samples amplify fine. I re-run these samples and they work fine. On failure, they produce a band of low molecular weight (100-200 bp) which sometimes looks like a smear with complete absence of my desired product size or my desired band is faint. For example, I will run 10 samples and 9 or 10 will work but one will fail. I am not using a hot start polymerase. However, my reaction mixture and reaction tubes are kept on ice at all times. I do pipette my mastermix up and down a 5-10 times before aliquoting which would be the only time other than aliqouting that the mastermix has a chance to briefly warm up. After adding gDNA to the reaction tube I do not mix, I just pipette the small volume of gDNA up and down a few times. I also have my thermal cycler pre-warmed.
Any explanations for the random failures?
Mastermix mixing problem? Should I vortex my mastermix lightly or pipette up and down before aliquoting?
Not mixing sample into reaction enough? Should I give them a slight vortex?
How sensitive is a non hot start polymerase to brief temperature increase (i..e few seconds during mixing or transferring to thermal cycler) ?
Thanks
I attached a picture. Top gel is what I am referring to and bottom gel too. Bottom gel also has an issue with the non-specific high MW bands. DNA ladder is 100bp with the brighest band = 500bp. The desired bands are supposed to vary in size because it is a genotype assay.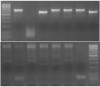
Hi Epigeneticist,
There could be a couple of reasons why your PCR fails for a few samples.
<*>SInce you are adding template DNA from various sources, their DNA concentration could be variable.
<*>If you are using lysis solutions for your extraction, salt remnants can also impact how well your PCR works out.
<*>Also, you said that you DO NOT mix your gDNA into your master mix. Well, I always mix it.
<*>How gDNA do you add? Porbably would be a good idea to get you pipette calibrated if you pipette volumes that are less than 2 ul
Hope this helps.
Ameya
Ameya P on Mon May 6 07:51:30 2013 said:
Hi Epigeneticist,
There could be a couple of reasons why your PCR fails for a few samples.
<*>SInce you are adding template DNA from various sources, their DNA concentration could be variable.
<*>If you are using lysis solutions for your extraction, salt remnants can also impact how well your PCR works out.
<*>Also, you said that you DO NOT mix your gDNA into your master mix. Well, I always mix it.
<*>How gDNA do you add? Porbably would be a good idea to get you pipette calibrated if you pipette volumes that are less than 2 ul
Hope this helps.
Ameya
Thanks for the response Ameya and your suggestions. My problem is due to the polymerase which is not a hot start enzyme. I was doing a poor job of keep my samples on ice during setup. I have been very careful to keep everything on ice and I haven't had any issues.
I ruled out the problems you suggested based on the following:
DNA concentration - I check my samples in duplicate and I even verified my diluted samples too.
Salt in DNA extractions - extractions are usually very good, and DNA eluates are concentrated so I have to dilute them which would dilute out the salt if I had any.
Pipettes calibration - calibrated by a certified company
Thanks again for your response! Do you have any explanation for the cause of non-specific high molecular bands (between 600-1000bp) or how to get rid of them? I have read too much template could cause this but I do not think 100ng of genomic DNA is excessive. My product is only 400bp so I decreased my extension time from 1min to 30 seconds and it got rid of the bands. Just wondering if there is another way to approach it.
