Uncut whole genomic DNA running too high - (Aug/08/2012 )
I am looking at DNA fragmentation in mammalian tissues. I extract whole genomic DNA from mouse tissue (skin, muscle, etc) by phenol-chloroform method. I am extremely careful not to break DNA during the extraction process. I digest with RNAse A for 30min at 37C before the last chloroform:IA extraction step. I then label 100ng of DNA with TdT and DIG-dUTP, run it out on a ~1% agarose gel, and do an anti-DIG southern blot.
The problem is that I get the expected smear, but the smear stops with a large fat band around 1.2kb (according to my ladder), then there is empty space, and then another fat band if my DNA had been intentionally fractionated or nothing if the DNA was not damaged.
My question is: what is this band at 1.2kb? Why doesn't the DNA run all the way down (I expect a continuation of a smear, getting lighter towards the bottom if it's undamaged whole DNA). Has anyone gotten this before?
Image legend: lane1: DNA damaged, lanes 2+3: DNA undamaged, lane 4: standard ladder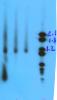
Truly intact genomic DNA won't come out of the well. It probably won't transfer to a southern blot either.
