RAPD - PCR problem - (Jul/07/2012 )
Hello everyone!
According to the Nanodrop Spectophotometer my dna sample of Holothuria tubulosa (sea cucumber) from gonads is pure.
The DNA extraction has been made with the CTAB Protocol.
The ratio is 1.91 and 2.31 for the 280/260 and 260/230. The concentration of my sample is 350 ng/ul.The curves and ratios are perfect so i think there is no problem with my samples according to nanodrop.
I have tried many (up to 35) PCR RAPD 10mer Primers with many combinations such as:
1) 2-5% DMSO
2) Different concentrations of DNA sample ( from 10-350 ng/ul )
3) Gradient PCR from 30-44C (annealing step)
4) Samples such as Holothuria tubulosa skin, Urchin and Shark (Urchin and sharks were amplified so i have positive samples.)
5) Different PCR steps such as long time denaturation (from 5 to 30 minutes), and long final extension step.
6) At 25μL and 30μL with different concentrations of Primer (0,3-3μL / 25μL), Buffer (2,5-10μL / 25μL) DNA (1-2μL / 25μL), DNTP (0,5-2,5μL/25μL) but i have no results.
Although Holothuria tubulosa gonads and skin has a problem in PCR amplification and i don't understand the reason why.
Any idea of what's preventing PCR amplification in that case?
Thank you!
Check your DNA on a gel - nanodrop only tells half the story.
It is very likely that there are some compounds such as glyco-proteins that have been extracted with the DNA.
First I thought of inhibitors that are still in the DNA (no idea if sea cucumbers have them) or that the DNA is too different for standard RAPD primers (if you used them). But as they're that short it's not very likely that there is no complimentary strand in the genome).
Thank you for responding!
Electrophoresis gel is not that good...it seems that DNA from gonads is degraded not in all samples (photo ''dna-extraction'').
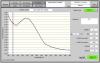
DNA from skin is perfect in gel but i have problems in nanodrop (see ''p9-Skin'' nanodrop photo).
As it seems there are some problems with my DNA but even the best samples cannot work in PCR.

Dear JohnSky,
To rule out inhibitors which might exist in your sample, you might consider to spike (or mix, as you call it) some working RAPD DNA in your RAPD PCR which is not sea cucumber origin (might be from plant, other organism) which is around 10%-20% of the total DNA concentration you had used in your PCR reaction.
I suggest you to do the following:
Saying if your total DNA concentration in for a successful run of "Plant" RAPD PCR is from 500 - 5ng/ul (random figure), in your "sea cucumber" RAPD PCR, try use a total genomic concentration of 100ng/ul in first tube, second tube you mix the PLANT DNA with SEA CUCUMBER in 1:9 ratio (so, for a 100ng genomic content 10ng is from PLANT), third tube just 10ng/ul PLANT DNA, fourth tube SEA CUCUMBER 90ng/ul. Run all your PCR together with your PCR profile. If you only got the third tube working, this means your inhibitor present. IF you got the third and second tube working and failed on the fourth tube, highly possible your RAPD primers is not working. IF your fourth tube is working, this might sounds that you need some dilutions in your template.
Just my 2 cents.
Well according to an article, the tissue from Holothuria tubulosa can not be preserved in ethanol neither in DMSO but only in liquid Nitrogen.
I got fresh samples and i am making new extraction and pcr. I hope i can take good results for my RAPD now...
Thank you for your help!
