shRNA oligo annealing problem - (Jan/27/2010 )
Hi all,
I am trying to generate an shRNA vector in the pLKO.1 backbone, and I'm getting stuck at the very first step: oligo annealing.
The oligo sequences are:
sense:
CCGGgtggatattgttgccatcaatCTCGAGattgatggcaacaatatccacTTTTTG
antisense:
AATTCAAAAAgtggatattgttgccatcaatCTCGAGattgatggcaacaatatccac
The capital letters indicate nucleotides that were recommended by the Addgene protocol for designing shRNA oligos for pLKO.1, and the lowercase letters are the sequence targeting my gene. I have five pairs of oligos, each differing in the targeting sequence.
I have tried three different annealing buffers, I tried the waterbath and the PCR cycler approach (95C for 4 min, 70C for 10 min, cool down to room temp over the course of several hours), and I can't get annealing to work.
I am checking the annealing reactions on a 3% agarose gel. No matter if I run the ss oligo, or if I run the (presumably) ds oligo after the annealing reaction, I see the same band pattern. (I should expect to see a slower-migrating band after annealing, right?)
Another weird thing is that the oligos themselves, without any annealing buffer or heating, show up as two bands on a 3% agarose gel. I am attaching a picture of the gel. You can see that for three sets of the oligos (#1-3), they run predominantly as an "upper" band, and for two sets (#4-5) they run as a "lower" band. I also tried combining the sense and antisense oligos (just in ddH2O, no annealing) and running the mixture on the same gel. You can see that the mixture also has a two-band pattern.
I am utterly confused as to what's going on. Any advice would be greatly appreciated.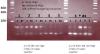
that is going to be my next step too, i'm not sure yet about the protocol completly but i think u should add T on your sense strand "CCGGT....." and on your antisense u should add A at the end of your seq. and that is because of the restriction enz u r about to use. if i rememebre corectly it's age1 and ecor1, so be aware of that
irrumator11 on Jan 27 2010, 01:53 PM said:
I am trying to generate an shRNA vector in the pLKO.1 backbone, and I'm getting stuck at the very first step: oligo annealing.
The oligo sequences are:
sense:
CCGGgtggatattgttgccatcaatCTCGAGattgatggcaacaatatccacTTTTTG
antisense:
AATTCAAAAAgtggatattgttgccatcaatCTCGAGattgatggcaacaatatccac
The capital letters indicate nucleotides that were recommended by the Addgene protocol for designing shRNA oligos for pLKO.1, and the lowercase letters are the sequence targeting my gene. I have five pairs of oligos, each differing in the targeting sequence.
I have tried three different annealing buffers, I tried the waterbath and the PCR cycler approach (95C for 4 min, 70C for 10 min, cool down to room temp over the course of several hours), and I can't get annealing to work.
I am checking the annealing reactions on a 3% agarose gel. No matter if I run the ss oligo, or if I run the (presumably) ds oligo after the annealing reaction, I see the same band pattern. (I should expect to see a slower-migrating band after annealing, right?)
Another weird thing is that the oligos themselves, without any annealing buffer or heating, show up as two bands on a 3% agarose gel. I am attaching a picture of the gel. You can see that for three sets of the oligos (#1-3), they run predominantly as an "upper" band, and for two sets (#4-5) they run as a "lower" band. I also tried combining the sense and antisense oligos (just in ddH2O, no annealing) and running the mixture on the same gel. You can see that the mixture also has a two-band pattern.
I am utterly confused as to what's going on. Any advice would be greatly appreciated.
ofira_carmi on Jan 27 2010, 03:45 PM said:
You're right; the cloning sites are AgeI and EcoRI. Having CCGG... at the one end of the shRNA creates the sticky end for AgeI ligation.
Here's what the Addgene protocol states:
To generate oligos for cloning into pLKO.1, insert your sense and antisense sequences from step B.1 into the oligos below. Do not change the ends; these bases are important for cloning the oligos into the pLKO.1 TRC-cloning vector.
Forward oligo:
5' CCGG—21bp sense—CTCGAG—21bp antisense—TTTTTG 3'
Reverse oligo:
5' AATTCAAAAA—21bp sense—CTCGAG—21bp antisense 3'
that's correct but the guy i got this system from told me that it's more efficient for the next step. that is from his own experience and he can't have a reasonble reason for that, it works for him. i haven't tried it yet, i guess i'll order my sh-oligos in the next week... i hope it will works for me ![]()
![]()
ofira_carmi on Thu Jan 28 17:45:52 2010 said:
that's correct but the guy i got this system from told me that it's more efficient for the next step. that is from his own experience and he can't have a reasonble reason for that, it works for him. i haven't tried it yet, i guess i'll order my sh-oligos in the next week... i hope it will works for me
HI Did your oligo annealing and cloning work fine? i Ma having trouble annealing the oligos to clone them in to PLko.1 vector cut with ageI and EcoRI. I also see a similar gel image where both the non annealed single stranded oligos show the same pattern as the others that were annealed.
cheers
thanks
Hi I am trying to clone my shRNA into pLKO.1 as well but have problems with the annealing. Was wondering if anyone had trouble with it and was finally able to get past this step successfully. Thanks!
