Troubleshooting ligation/transformation - strange results when troubleshooting (Dec/09/2009 )
Hello all friendly cloners.
My student and I have been trying desperately to complete what I thought was going to be a simple cloning task. We are trying to insert a <4kb PCR fragment into a lentiviral vector using the enzymes EcoRI and Xba.
1. PCR with RE primers and column purify (Qiagen) confirm by running few ul on a gel -its always the correct size.
2. Set up sequential digests since NEB recommends different buffers for EcoRI and Xba. we column purify after each and confirm we have not lost any DNA by running a few ul on a gel afterwards.
3. In the last 30 min of the second digest (XbaI) we a 1 ul of CIP to the vector only.
4. So far the vector and PCR fragments have been confirmed to be present and the right size at each step.
5. we have then set up various ligations using 2 different ligases that I had in the freezer. We've tried overnight and few hours at RT.
6. we have then tried transformation of 1-2 ul into either electrocomp. cells or chem comp max efficiency cells and have gotten NO colonies on several different attempts (both with and without insert added in ligation). We know that the plates are OK because we recently transformed the empty vector onto these same AMP plates.
7. After reading some forums I set up a simple ligation with the PCR fragment only to see if I would get a band that was double the original size to show the ligase is active. When I did this I got a very strange result. I got a faint band at the original size of the fragment and I got a SMALLER band at about 1 kb! what could this be? It was not there before the ligation (I rechecked all of our images).
At this point I still don't know if our ligases are working (I even tried some of one I stole from another nearby lab) or what else is going on.
Please help. The attached picture is the recent ligation with 3 different ligases. Also I have never seen a smear like that before!
![]()
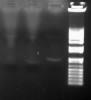
Did you heat kill the ligase before running the gel? Otherwise, the ligase will bind the DNA and mess up the gel.
Things to immediately check:
* Are there overhangs on the PCR product primers to allow the restriction enzymes to cut
* Try the ligation and transformation without CIP
* Test the transformation efficiency of your cells/protocol. You should be able to get 20-100 colonies from 10 pg of your uncut vector.
