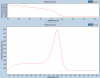
Melt curve analysis - (Nov/12/2012 )
Dear Guys,
I tried to read more to understand Rt-PCR, to perform a good reliable experiment.
I read about melt curve and it helps u to identify unspecific primer-dimer.
and I saw an example.
but let me say my question according to the example,
Now I generated my melt curve, it will show decrease in fluorescence as function of temp.
so if I have 2 peaks??? according to definition of Tm, it is temp which 50% of dsDNA bases have been denatured??
so how I can know which one is my true Tm.
or just finding 2 peaks is enough to say, I have unspecific product, lets repeat the experiment in another way???
I am very grateful for those who is welling to help me with their experience
Best Regards
Madeline
Not having done this for a while (years) I may be mis-remembering, but 2 peaks indicate the presence of 2 products, which is definitely what you do not want - you'll probably have to re-design primers.
The "peaks" are negative derivations, you should be able to see even the underived curve somewhere in program, which in case of two peaks will have one decrease in fluorescence in the area of first, and second in the area of the second peak.
Generally two peaks are bad if there should be one specific product, it could be either primer-dimers or unspecific product. Tm (the peak maximum) of the primer-dimers is usually substantially lower than that of product.
You can get rid of dimers or unspecifities by program optimisation, but not always, dimers are usually more difficult.
Post a picture and we can analyse it in detail.
Dear
Trof
Thanks for ur reply, I have not performed my experiment yet, I am just studying more about Rt-PCR and its optimization procedure, so I could not understand this point especially when I have never saw melt curve in real life except in drew in papers.
I wanted to send you some of my melting curves;
1. one positive sample with a single peak;
2. one NTC (which has no peaks);
3. one with the specific product and primer-dimers (which are smaller as Trof says and are located on the left side);
4. one with two products (two peaks, both located on the right side)
but I am not allowed to upload emf files.
Here I have a veeeeeeeery old PC and even don`t know with what kind of program to open them.
Never mind.
I don`t know what we should do if we have variant 3 in our results (maybe just optimization) but I know that variant 4 is the last thing that we want to see when we do qPCR :-)))))))))))
I think, this should mean either contamination (because primers are not very specific) or two specific products which result from splicing variants. This is what I`ve been told.
When I started to introduce myself with the real-time Trof gave me the Best Advise I Have Ever Got, which was ''Just start doing your real-time experiments and see what will happen. No reason to worry about the problem before you have it.''. So simple and effective.
I hope this will help also you. We can not become experienced if we do not gain experience :-)
Good luck :-)
I made some (though it was pretty tough to find any dimers ![]() )
)
The higher one is a graph of normalised fluorescence during melting, the lower one is second derivation of that curve.
First an example of specific product, two different specific product (different genes in the same run). The peak of the derivative curve is Tm (50 % of product melted or local inflection point).
The product with the higher Tm is a classic example of fine specific product, the one with lower Tm is also free of dimers, but the peak is bit wider, which is probably still OK, but sharper the better.

Next are examples of unspecific products.
The first case has a small peak with higer Tm than the main product, so that quite sure a nonspecificity (non-specific binding of primers to template), not a dimer.
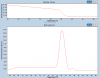
The other one has nonspecific product with lower Tm, but it's pretty abundant, so it's most likely also non-specific binding.

Now the dimers. Dimers are product created by binding of primers together, so the created product is usually pretty short, so Tm is much lower. Peaks with much lower Tm are therefore most likely dimers.
First one has a pretty small but still noticable dimer.

The other one more distinct one.
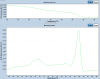
And at the end some tricky one, a single yet wide peak can also appear.
Actually there is a hidden second product with very similar Tm hidden in the curve, so also not good.